Lung Cancer
Lung Cancer
Lung cancer is the deadliest cancer worldwide (American Cancer Society, 2020). Diagnosis requires highly specialized physicians and is prone to errors due to the hard-to-see nodules, from which nearly 35% are missed in initial screenings (E. Svoboda, 2020). The success of lung cancer treatment and mortality reduction is directly related to the stage where cancer is diagnosed. The earlier it is diagnosed, the higher chances of survival. Unfortunately, lung cancer is usually diagnosed in the fourth stage. This problem motivates our work to improve CAD methods for early lung cancer diagnosis.
Motivation
Multimodal Analysis
Even though previous methods use multimodal information, they do not consider the anatomical spatial elationship familiar to radiologists, such as the existing correlation between finding nodules in multiple locations and the cancer stage. We believe the LUng CAncer Screening with Multimodal Biomarkers (LUCAS) (Daza et al., 2020) dataset is a promising experimentalframework, for it is the first that provides clinical information considered by physicians in addition to chest CT scans.
In this paper we propose a method that exploits multimodal data by spatially combining visual and clinical information to predict a patient’s probability of having lung cancer.
Method
Method
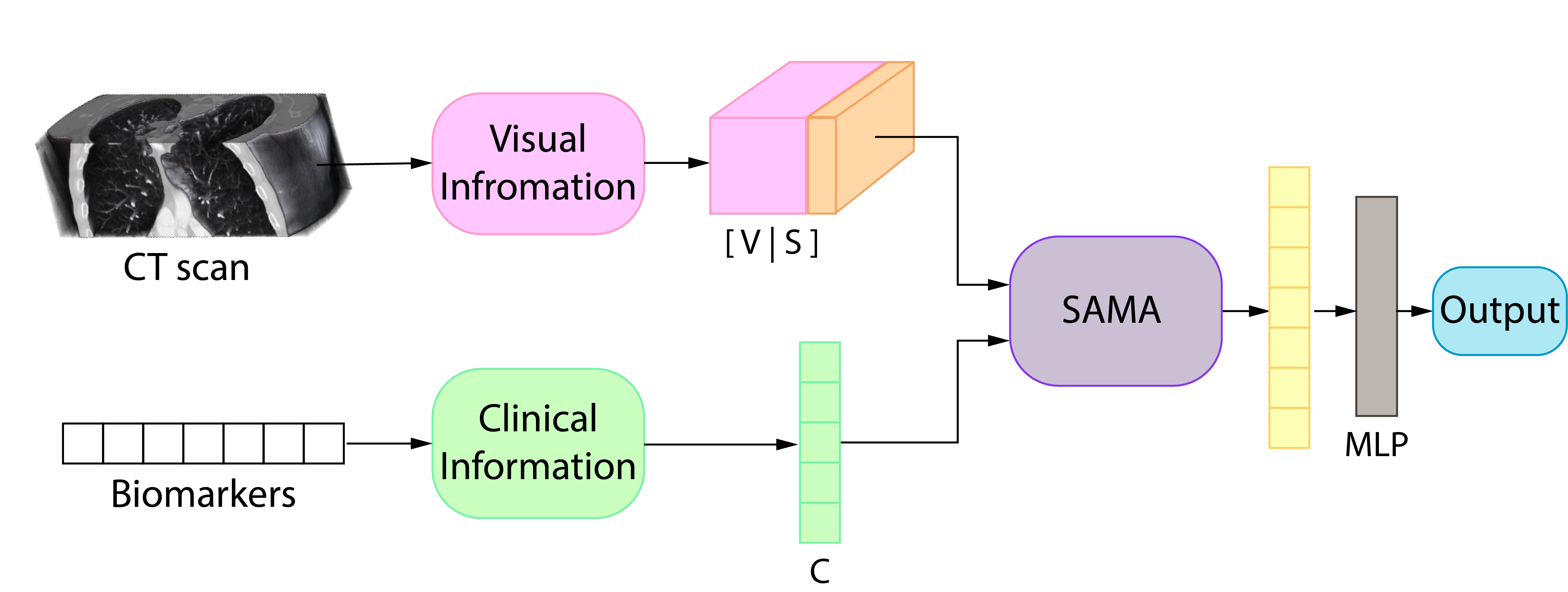
- Our method processes information in two parallel branches, one for each data modality.
- For the visual information, we use the backbone proposed by Daza et al. producing a visual representation (V).
- The clinical information goes through an MLP outputting a clinical representation (C).
- We process the two branches’ output using our SAMA module
Spatially Aware Attention
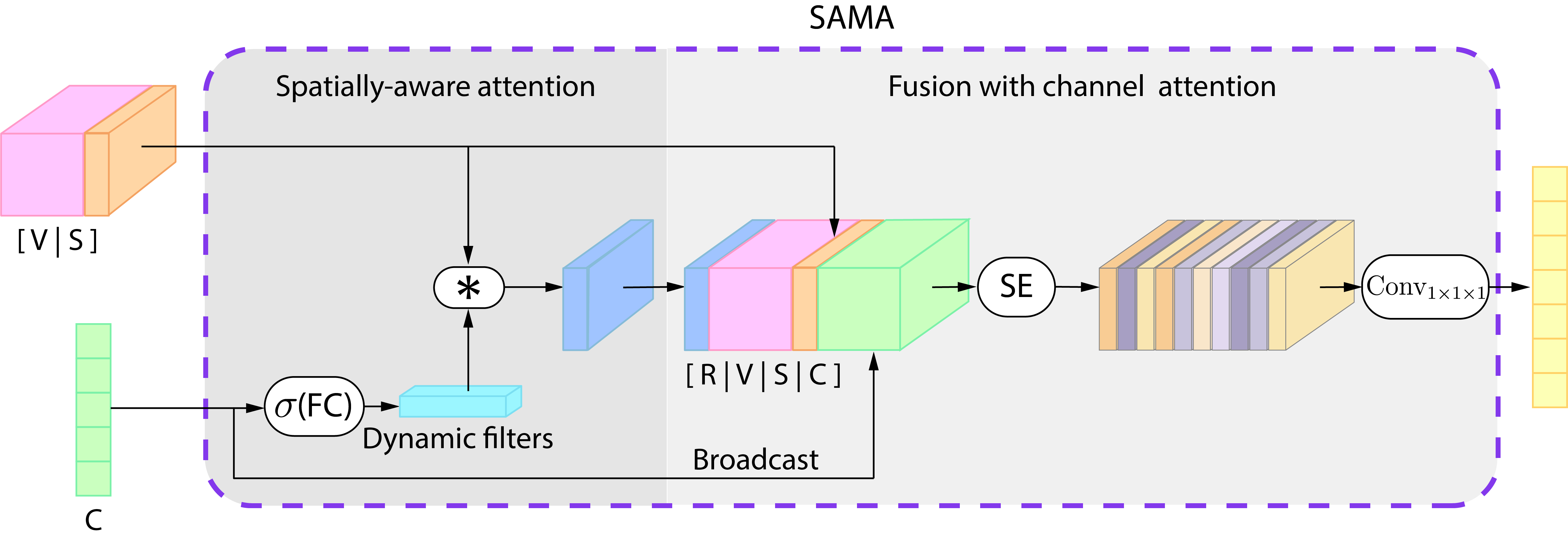
- We generate dynamic filters (Li et.al, 2017) using a dynamic convolutional layer, which produces new filters depending on the patients’ clinical information (embedded in the clinical representation).

- We generate a 3D spatial coordinate representation (S) of the visual information
- We calculate a dynamic response map R as:

Fusion with Channel Attention
- We concatenate response maps, visual, spatial and clinical data
- This concatenation goes through a Squeeze-and-Excitation block (SE) (J. Hu, et al., 2018)
- This weighted representation goes through a pointwise convolutional layer to fuse all the multimodal information at each location independently, producing the fused representation:

- Finally, the output goes through a Multilayer perceptron (MLP) with one hidden layer
- The network outputs a predicted score representing the probability of having cancer
Results
Results
- SAMA outperforms the state of the art by 14% in F-1 score and 16% in AP:

- Multimodal data enhances the model’s performance:
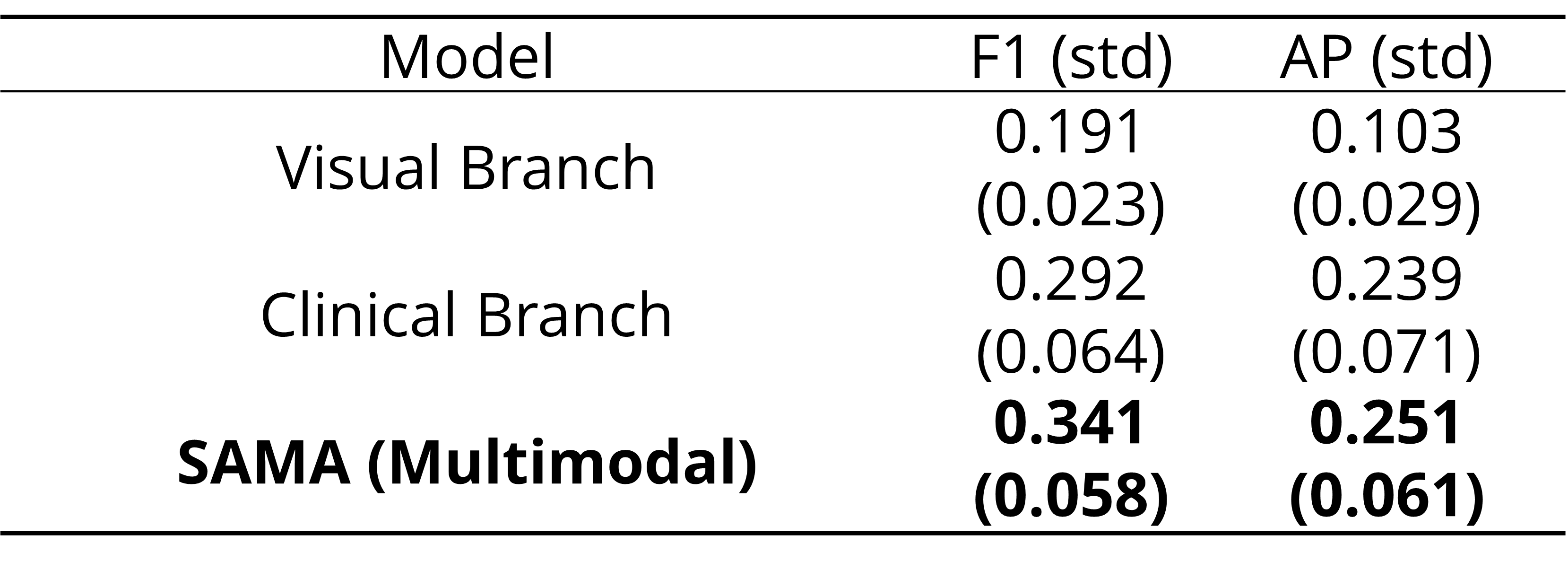
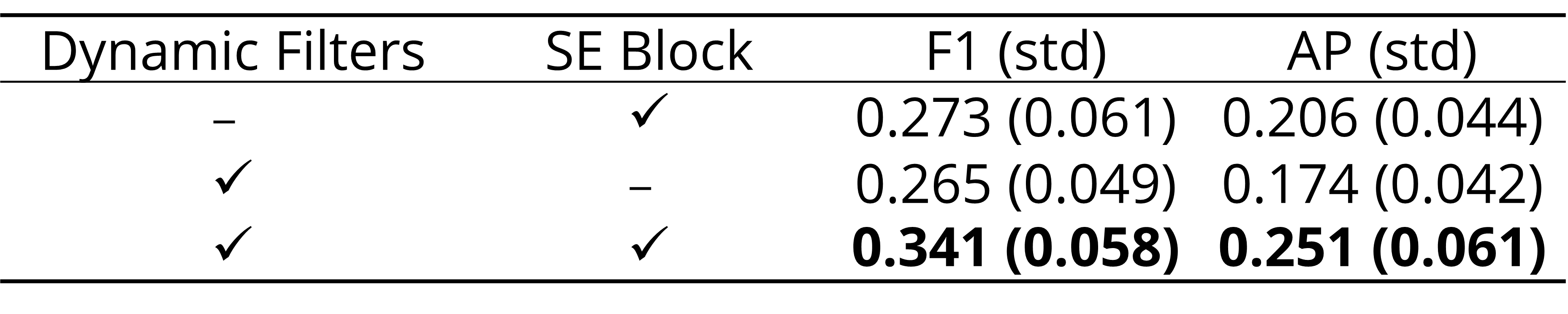
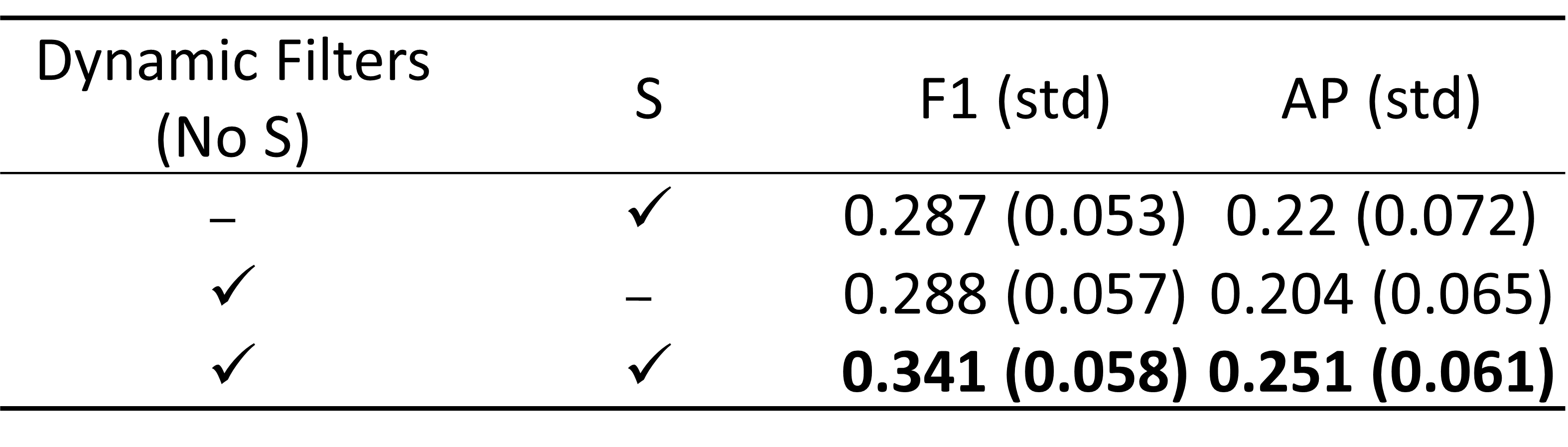
- All of SAMA’s components are crucial and improve performance:
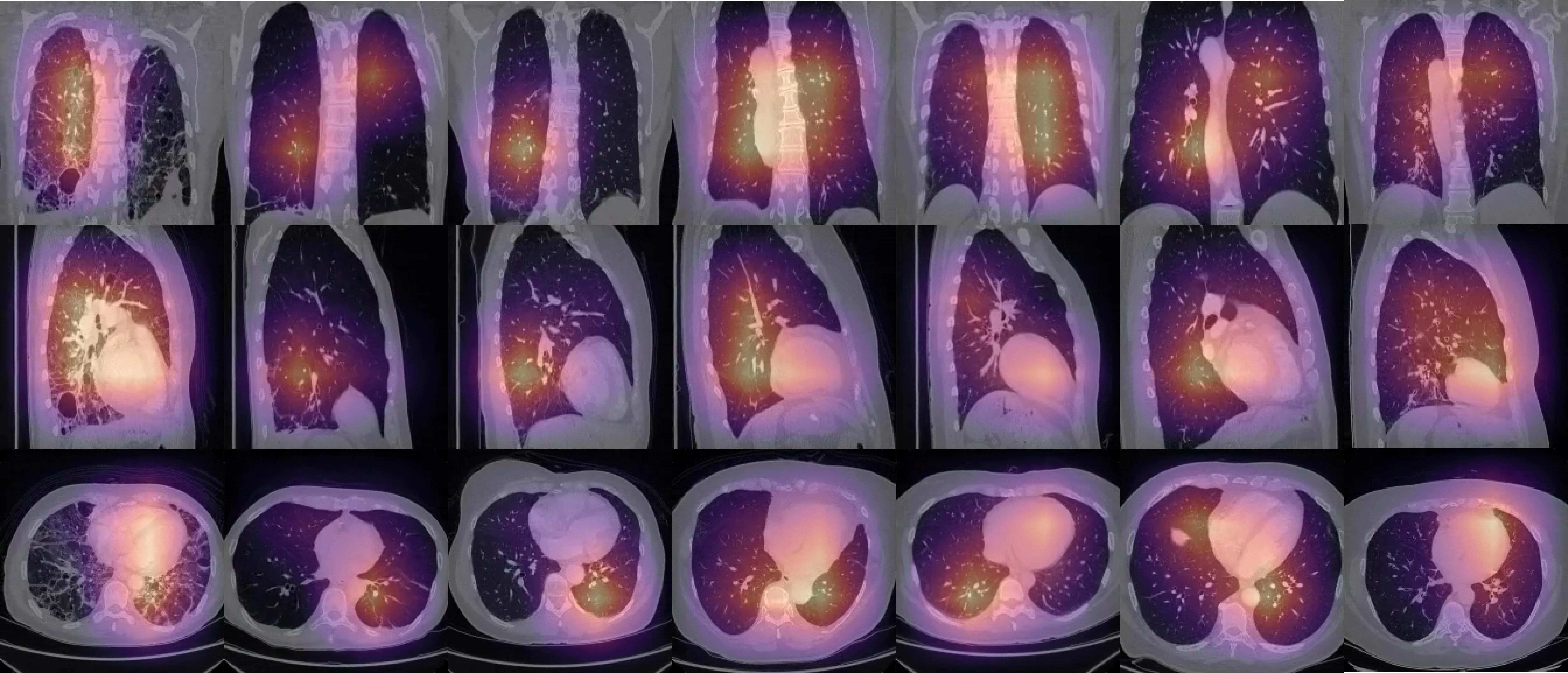
Qualitative Results
SAMA’s attention is focused on most relevant locations in an unsupervised manner: